Uncovering the drivers of seasonal respiratory infection dynamics |
|---|
|
Fundings: DMM at the University of Padova, the program Starting Package from the Cariparo Foundation, the program Emergence(s) of municipality of Paris. Selected pre-print and publications: F Bonacina, P-Y Boëlle, V Colizza, O Lopez, M Thomas, C Poletto, medRxiv (link) F Bonacina, P-Y Boëlle, V Colizza, O Lopez, M Thomas, C Poletto, Int J Inf Dis 128 (2023) (link) Recent master thesis: Ioannis Krimitsas, Hidden pathogen-pathogen interactions in a multi-pathogen ensemble, master in Physics, University of Padova (link) Marika Sartore, Dynamics of interdependent epidemics, master in Physics of Data, University of Padova (link) Forecasting activity: We are contributing to the Influcast Hub run by ISI Foundation. Together with other research groups we are providing each week model forecast of the number of cases of Acute-Respiratory-Infections and influenza. |
Unraveling the complex dynamics of multiple pathogens/strains |
|---|
|
Fundings: The program Emergence(s) of municipality of Paris. Selected pre-print and publications: P Toranj Simin, JC Taube, E Vergu, S Bansal, L Opatowski, C Poletto medRxiv (link) F Pinotti, É Fleury, D Guillemot, P-Y Böelle, C Poletto PLoS Comput Biol 15(5) (2019) (link) C Poletto, S Meloni, V Colizza, Y Moreno, A Vespignani, PLoS Comput Biol 9(8): e1003169 (2013) (link) |
Outbreak analysis |
|---|
|
Fundings: Haute Autorité de Santé, ANR Flash Covid-19, the Fondation de France, the program Emergence(s) of municipality of Paris. Selected publications: B Faucher, CE Sabbatini, P Czuppon, MUG Kraemer, P Lemey, et al Nat Commun 15, 2152 (2024) (link) JA Moreno López, B Arregui-Garcĺa, P Bentkowski, L Bioglio, F Pinotti, et al Sci Adv 7 (2021) (link) F Pinotti, L Di Domenico, E Ortega, M Mancastroppa, G Pullano, et al PLoS Med 17(7) (2020) (link) J Riou, C Poletto, P-Y Boëlle, Epidemics 19 (2017) (link) C Poletto, C Pelat, D Levy-Bruhl, Y Yazdanpanah, P-Y Boëlle, et al Euro Surveill 19:23 (2014) (link) C Poletto, M F C Gomes, A Pastore y Piontti, L Rossi, L Bioglio, et al Euro Surveill 19:42 (2014) (link) D Balcan, H Hu, B Goncalves, P Bajardi, C Poletto, et al BMC Med, 7:45 (2009) (link) COVID-19 real-time analyses: Evolution du nombre de tests positifs, des nouvelles hospitalisations et des admissions en soins critiques pour COVID-19 apres la mise en place des mesures sanitaires renforcées, Mars-Avril 2021 (available here) Evaluation des stratégies vaccinales COVID-19 avec un modèle mathématique populationnel (available here) Doubling time of COVID-19 hospitalizations in regions in France (available here) Evaluation of StopCovid (available here) |
Conceptual tools for outbreak analysis |
|---|
|
Selected pre-print and publications: PK Kollepara, C Poletto, JC Miller, medRxiv (2025) (link) E Valdano, D Colombi, C Poletto, V Colizza, Nat Commun 120, 068302 (2023) (link) A Apolloni, C Poletto, J J Ramasco , P Jensen, V Colizza, Theor Biol Med Model 11 (1), 3 (2014) (link) Recent master thesis: Lorenzo Luzzi, Biased epidemic analysis due to heterogenous and partial surveillance: a study on deterministic and stochastic multi-class models for epidemic dynamics, master in Mathematics, University of Padova (link) |
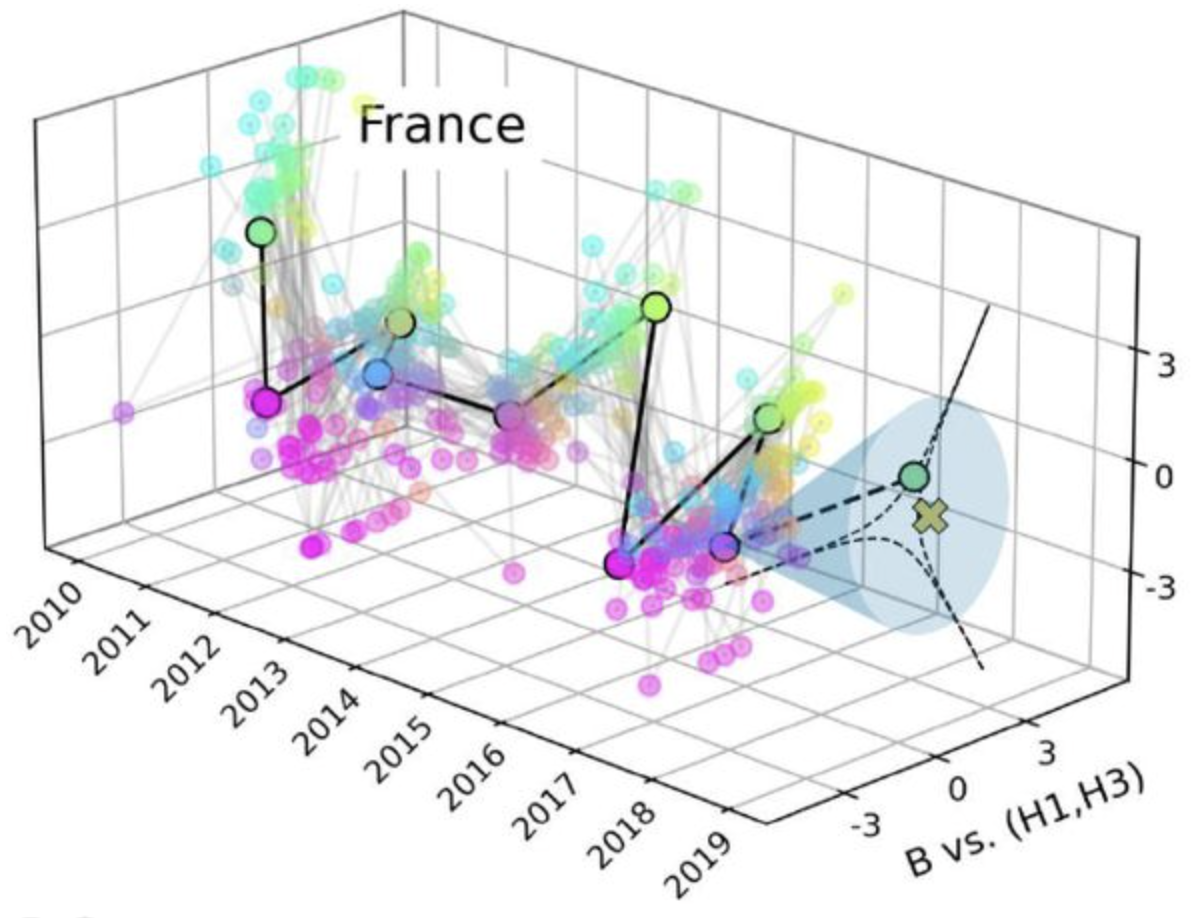 Each winter, seasonal respiratory viruses cause a substantial number of infections,
hospitalizations, and deaths. Among these pathogens, influenza represents a major source of public health burden. Despite decades of research,
many questions remain open regarding its transmission and evolution: how does the interplay between social contact
patterns and age-specific susceptibility shape epidemic dynamics? How do different viral variants interact, and how
can we explain their co-circulation and coexistence across spatial and temporal scales? Beyond influenza, a multitude
of respiratory viruses co-circulate and interact, some of which impose a disease burden comparable to that of influenza,
such as respiratory syncytial virus (RSV). Understanding and predicting the dynamics of this complex ecological
systems is a key research and public health priority, requiring complex-systems thinking and advanced physical,
statistical, and computational approaches.
Each winter, seasonal respiratory viruses cause a substantial number of infections,
hospitalizations, and deaths. Among these pathogens, influenza represents a major source of public health burden. Despite decades of research,
many questions remain open regarding its transmission and evolution: how does the interplay between social contact
patterns and age-specific susceptibility shape epidemic dynamics? How do different viral variants interact, and how
can we explain their co-circulation and coexistence across spatial and temporal scales? Beyond influenza, a multitude
of respiratory viruses co-circulate and interact, some of which impose a disease burden comparable to that of influenza,
such as respiratory syncytial virus (RSV). Understanding and predicting the dynamics of this complex ecological
systems is a key research and public health priority, requiring complex-systems thinking and advanced physical,
statistical, and computational approaches.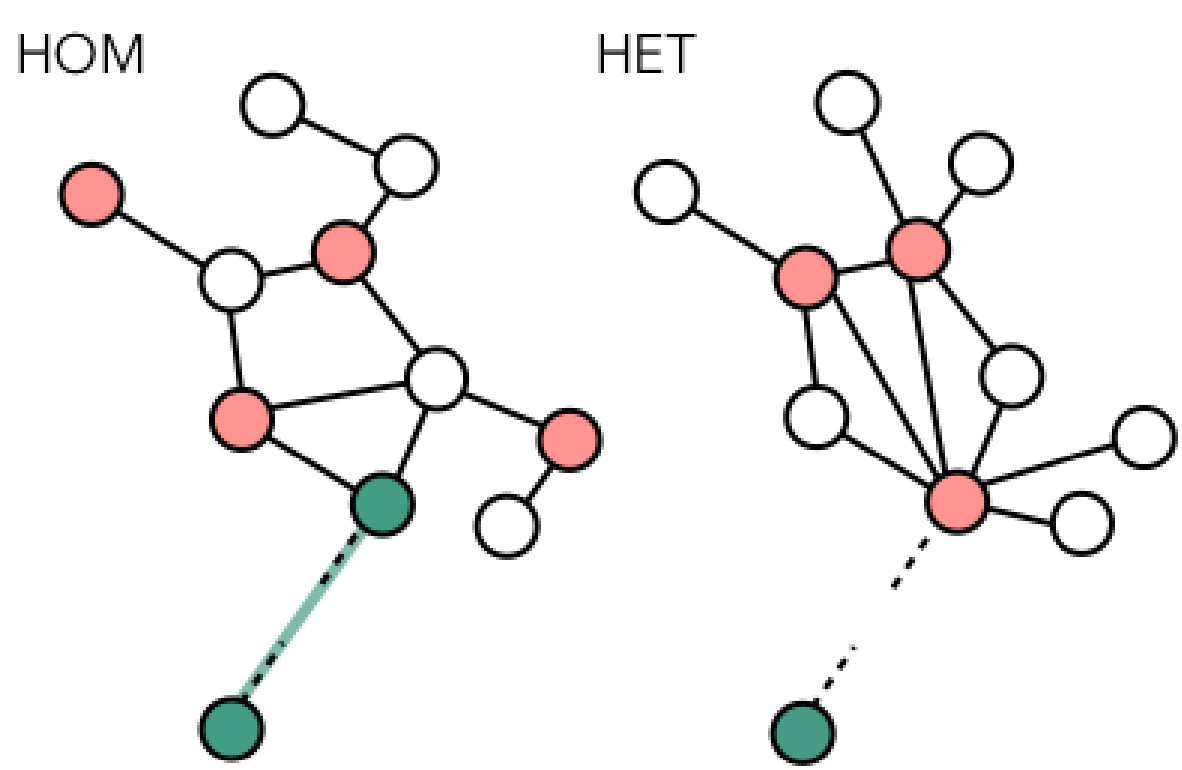 Across biology, ecology, epidemiology, and public health, increasing attention is being
devoted to the web of interactions among multiple infectious agents - either different
pathogens or different strains of the same pathogen - and between pathogens and their hosts.
Pathogens do not spread in isolation; rather, disease outbreaks emerge from complex ecological
processes in which infectious agents compete, cooperate, and evolve. The human contact network,
which provides the substrate for transmission, is heterogeneous, high-dimensional, and dynamic.
Understanding the interplay between host behavior and multi-pathogen or multi-strain interactions
is crucial for addressing major public health challenges, such as the spread of antibiotic resistance
and the co-circulation of seasonal respiratory viruses. Alongside empirical studies of co-circulation
patterns, we develop mathematical and computational models to uncover the dynamical principles
governing these complex systems. Our work contributes to disease ecology and addresses
open problems in network physics.
Across biology, ecology, epidemiology, and public health, increasing attention is being
devoted to the web of interactions among multiple infectious agents - either different
pathogens or different strains of the same pathogen - and between pathogens and their hosts.
Pathogens do not spread in isolation; rather, disease outbreaks emerge from complex ecological
processes in which infectious agents compete, cooperate, and evolve. The human contact network,
which provides the substrate for transmission, is heterogeneous, high-dimensional, and dynamic.
Understanding the interplay between host behavior and multi-pathogen or multi-strain interactions
is crucial for addressing major public health challenges, such as the spread of antibiotic resistance
and the co-circulation of seasonal respiratory viruses. Alongside empirical studies of co-circulation
patterns, we develop mathematical and computational models to uncover the dynamical principles
governing these complex systems. Our work contributes to disease ecology and addresses
open problems in network physics.
 (Re-)emerging pathogens pose a major public health threat. In particular, when a pathogen spills over from animal
reservoirs into humans or when known pathogens evolve, biological and epidemiological uncertainties can hinder a
timely response. Addressing this challenge requires improving our ability to understand outbreak dynamics through
the integration of data analysis, computational methods, and epidemiological modeling. We have conducted real-time
analyses to assess epidemic risk, generate projections, and quantify the impact of potential interventions, with
applications to major outbreaks including the 2009 A/H1N1 influenza pandemic, the emergence of MERS-CoV in 2012, the
2014 West African Ebola outbreak, Zike widespread in 2016, and the COVID-19 pandemic. For the latter, we provided
real-time analyses of epidemic spread - particularly focusing on the risk of international propagation - and
developed agent-based models to evaluate interventions such as digital contact tracing and reactive vaccination.
These studies were communicated to, or conducted in direct collaboration with, public health authorities to support
decision-making. In non-emergency periods, we continue carrying out retrospective analyses to extract lessons learnt
for future outbreaks.
(Re-)emerging pathogens pose a major public health threat. In particular, when a pathogen spills over from animal
reservoirs into humans or when known pathogens evolve, biological and epidemiological uncertainties can hinder a
timely response. Addressing this challenge requires improving our ability to understand outbreak dynamics through
the integration of data analysis, computational methods, and epidemiological modeling. We have conducted real-time
analyses to assess epidemic risk, generate projections, and quantify the impact of potential interventions, with
applications to major outbreaks including the 2009 A/H1N1 influenza pandemic, the emergence of MERS-CoV in 2012, the
2014 West African Ebola outbreak, Zike widespread in 2016, and the COVID-19 pandemic. For the latter, we provided
real-time analyses of epidemic spread - particularly focusing on the risk of international propagation - and
developed agent-based models to evaluate interventions such as digital contact tracing and reactive vaccination.
These studies were communicated to, or conducted in direct collaboration with, public health authorities to support
decision-making. In non-emergency periods, we continue carrying out retrospective analyses to extract lessons learnt
for future outbreaks. 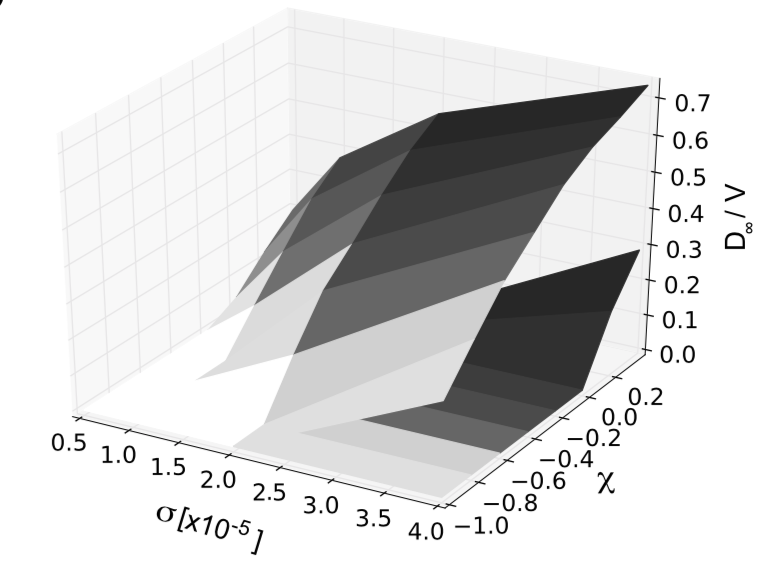 To understand how disease outbreaks develop, we rely on mathematical tools that help
us make sense of data, uncover hidden patterns, and reveal processes that cannot be
directly observed. These tools allow us to address key questions: under which conditions
does a small number of cases turn into a large outbreak? How do contact patterns shape
the characteristic timescales of transmission and epidemic spread? How does the
observation process - such as data collection and reporting - distort the apparent epidemic
trajectory, and how can we reconstruct the underlying dynamics? By developing and refining
these approaches outside of emergency situations, and drawing on ideas from mathematics
and physics, we can be better prepared to interpret data rapidly and respond effectively
when the next outbreak occurs.
To understand how disease outbreaks develop, we rely on mathematical tools that help
us make sense of data, uncover hidden patterns, and reveal processes that cannot be
directly observed. These tools allow us to address key questions: under which conditions
does a small number of cases turn into a large outbreak? How do contact patterns shape
the characteristic timescales of transmission and epidemic spread? How does the
observation process - such as data collection and reporting - distort the apparent epidemic
trajectory, and how can we reconstruct the underlying dynamics? By developing and refining
these approaches outside of emergency situations, and drawing on ideas from mathematics
and physics, we can be better prepared to interpret data rapidly and respond effectively
when the next outbreak occurs.